Evaluation Guide for Real-Time PCR Master Mixes
When evaluating a new real-time PCR master mix, it's essential to consider the right parameters. This guide provides a simple protocol and tips to help you quickly assess the performance of a real-time PCR master mix.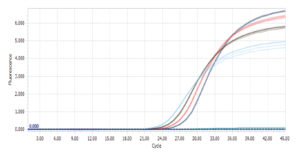
Amplification plot for RealQ Plus Green run on LightCycler® 96
1. Prepare a DNA Dilution Series
Create a series of DNA dilutions based on the diagram provided. Ensure the tubes are mixed thoroughly by gently flicking them (do **not vortex**!) and then spin them down. 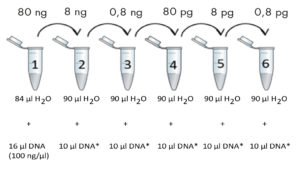 For genomic DNA, include tubes 1-4. For other types of DNA, include tubes 1-6. The initial DNA concentration is 100 ng/µl. The final DNA concentrations in each tube are indicated in the boxes above the tubes.2. Prepare a Reaction Mix
Mix the reaction components according to the table below. After adding all the components, briefly vortex for 2 seconds and then spin down. 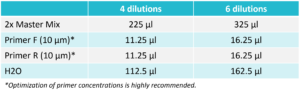 For Probe: 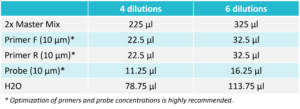3. Distribute the Reaction Mix
Distribute 20 µl of the reaction mix into the bottom of the blue wells as shown in the diagram below. 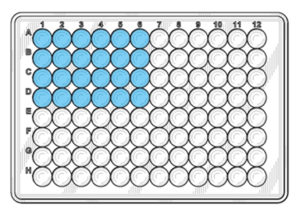4. Distribute the DNA
Add 5 µl of DNA from the dilution series into the respective wells. Ensure the pipette tip is submerged as little as possible into both the liquid of the tube and the well. The DNA should be distributed into the liquid and not onto the sides of the wells. 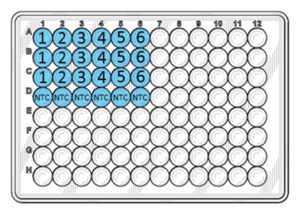 *Note: Do not pipet up and down into the wells; the DNA will mix during the initial heating of the PCR run.* The numbers in the figure above correspond to the tube numbers in the dilution series from step 1. Add PCR Grade Water to the NTC wells instead of DNA.5. Run the Plate
Use the following PCR cycling protocol when running the plate on the real-time PCR instrument. Run the melt curve according to instrument default settings. 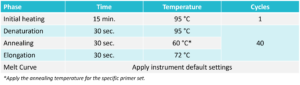 For Probe: No Melt Curve settings6. Evaluate the Results
The results should meet the specifications listed below to accept the mix. Use the table below to evaluate the mix. 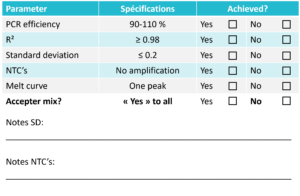 | Standard Curve | Melt Curve | |----------------|------------| | [Image](http://bsg-i.nbxc.com/blog/c152758efcba06296a011e48d66ef6f4.jpg) | [Image](http://bsg-i.nbxc.com/blog/1e6ecb39e4e2fe4bff03ff40731d1a13.jpg) | For Probe: No Melt Curve resultNotes
The results should fall within the specified parameters to accept the mix. Keep in mind that the melt point and Cq-values are buffer-dependent and may vary slightly between experiments conducted with different buffers. For a deeper understanding of what to look for when evaluating a mix, refer to the guidelines below:Standard Curve:
When analyzing the standard curve, focus on three key parameters: PCR efficiency, R²-value, and standard deviation between replicates. The PCR efficiency indicates how efficiently the target has been amplified and should range between 90-110% to be considered acceptable. This is one of the most critical parameters when evaluating a mix. The R²-value is a statistical measure indicating how closely the data fit the regression line and should be ≥ 0.98. The standard deviation between replicates reflects the accuracy of pipetting. This value should be ≤ 0.2.Melt Curve:
When examining the melt curve, there should be one distinct peak at the intended melt temperature of the product. It's important to note that the melt temperature is pH- and buffer-dependent. Therefore, it's not expected that PCR products from two different mixes will have identical melt temperatures. A slight variation in melt temperature is not indicative of how well the mix performs. The NTCs should not exhibit a peak at the same melt temperature as the intended product. If this occurs, there may be contamination, which could lead to false positive results. Other peaks besides those from the intended product can appear in the melt curve plot and indicate non-specific amplification. As long as these peaks are very small or the NTC wells appear on the amplification plot ≥ 40 cycles or ≥ 10 cycles after the lowest DNA concentration, the non-specific amplification can be disregarded.Cq-value:
Cq-values should be evaluated in relation to PCR efficiency and not be the sole focus of interpretation, especially as long as PCR efficiency remains within the acceptable range (90-110%). An extremely high Cq-value can be due to low PCR efficiency, which might be detected during the evaluation of efficiency. Inconsistencies in Cq-values can also stem from inhibitors in the sample and may result in an efficiency greater than 110%. Moreover, Cq-value can be influenced by SYBR levels, ROX levels, and target length (in SYBR experiments), leading to variations between master mixes. Minor differences in Cq-values are expected when comparing two mixes.SYBR Level:
It's less relevant to focus on SYBR levels when evaluating a master mix. These levels can be adjusted by simply adding more SYBR to the buffer and may not necessarily reflect the performance of the mix. Excessive SYBR can inhibit PCR amplification, but this would be detected when assessing PCR efficiency. Additionally, SYBR levels can be affected by ROX if the amplification plot is plotted with Rn or ΔRn on the y-axis. Only in the multicomponent view of an amplification does ROX not influence the Cq-value.Comparing Two Mixes:
If comparing two different master mixes, prepare a reaction mix with the other master mix as well, maintaining the same primer concentrations. If the same PCR cycling protocol is used, distribute the reaction mix and DNA dilutions in wells A7 to D12 as per steps 3 and 4. Run the melt curve according to instrument default settings. If the other master mix uses a different PCR cycling protocol, distribute the reaction mix and DNA dilutions into a second PCR plate as per steps 3 and 4. Run the plates separately. Run the melt curve according to instrument default settings.Other Parameters:
This guide emphasizes simplicity and minimizing required work time. However, for a comprehensive evaluation of a master mix, additional parameters should be considered. Key factors include: - Primer and probe design - Annealing temperature - Primer and probe concentration - Sample concentration - Inhibitor contamination - Choice of controls - Setting thresholds - Correct fluorescence chemistry Considering all these parameters allows for a thorough evaluation, ensuring the experiment setup is optimized.Extract from Ampliqon Items:
- RealQ Plus 2x Master Mix Green Low ROXâ„¢ A324499 - RealQ Plus 2x Master Mix for Probe Low ROXâ„¢ A314499Learn More:
- Flyers: - Study of Fast Cycling Real-Time PCR Conditions using RealQ Plus Master Mixes - REALQ PLUS MASTER MIXES - Search Real Time Master Mixes online - Contact us for support: - Phone: +33 4 70 03 73 06 - Email: This guide offers a straightforward approach to evaluating real-time PCR master mixes. By focusing on the essential parameters, you can ensure your PCR reactions are accurate and reliable.Power Wheelchair Accessories,Power Chair Seat Cushions,Wheel Chair Headrest,Cell Phone Holder For Wheelchair
Suzhou Danyazhihe Technology Co., Ltd , https://www.autowheelchairs.com